
Tutorials

1. Overview
The aim of RADB is to effectively integrate and analyze all the RA-related genetic polymorphisms extracted from published papers. We searched the PubMed database (http://www.ncbi.nlm.nih.gov/pubmed) using the following keywords: ((polymorphism[Title/Abstract] OR polymorphisms[Title/Abstract] OR GWAS[Title/Abstract] OR GWA[Title/Abstract]) AND rheumatoid arthritis[Title/Abstract]) NOT review[Publication Type]
As a result, we obtained about 1,500 publications. After manually scanning the list, 522 studies were included in RADB, comprising nine candidate gene linkage analysis studies, 505 candidate gene association studies, and eight genome-wide association (GWA) studies. We extracted the important information from these reports, including basic information about the article (e.g., PubMed ID (PMID), title, and abstract), population information (e.g., race and sample size), and polymorphism information (e.g., polymorphism name, host gene, genotype, odds ratio (OR) with 95% confidence interval (CI), P-value, and risk allele).
2. Abbreviations list
| No. | Full name | Abbreviation |
|---|---|---|
| 1. | Anti-cyclic citrullinated peptide antibodies | ACCP |
| 2. | Anti-tumor necrosis factor | Anti-TNF |
| 3. | Cardiovascular | CV |
| 4. | C-reactive protein | CRP |
| 5. | Denotes disease activity score | DAS28 score |
| 6. | Disease-modifying antirheumatic drugs | DMARDs |
| 7. | Erythrocyte sedimentation rate | ESR |
| 8. | Elderly-onset rheumatoid arthritis | EORA |
| 9. | Health assessment questionnaire | HAQ |
| 10. | Healthy controls | H |
| 11. | Osteoarthritis | OA |
| 12. | Odds ratio | OR |
| 13. | Rheumatoid arthritis | RA |
| 14. | Rheumatoid factor | RF |
| 15. | Single nucleotide polymorphism | SNP |
| 16. | Visual analog scale | VAS |
3. Data categories
According to the types of case-control and research purposes, data is divided into four categories.
(i) Susceptibility locus for RA
People who carry these alleles have a higher risk of developing rheumatoid arthritis than those without these alleles.
(ii) Polymorphisms associated with the clinical features of RA
Clinical features of RA, such as rheumatoid factor (RF) status, Anti-citrullinated peptide antibodies (anti-CCP) status and age of onset, are often associated with genetic factors.
(iii) Polymorphisms affected drug treatment outcome in RA
Disease-modifying antirheumatic drugs (DMARDs) and anti-tumor necrosis factor (anti-TNF) agents are the mainstay of treatment in rheumatoid arthritis. However, inconsistent response to these drugs is often observed, with considerable variability in both efficacy and toxicity. It is possible that genetic component might influence drug treatment outcome in patients with RA.
(iv) Polymorphisms associated with a higher risk of cardiovascular disease in RA
Rheumatoid arthritis is associated with an increased risk of cardiovascular (CV) events, causing an increased cardiovascular (CV) morbidity and mortality. It is supported that the development of CV disease in RA is related to genetic component.
4. Search
Different users may have different needs, so we designed a variety of ways to search RADB. In addition to queries, users can also list all of the polymorphisms, all of the genes or all of the literatures.
4.1 Search by polymorphism name
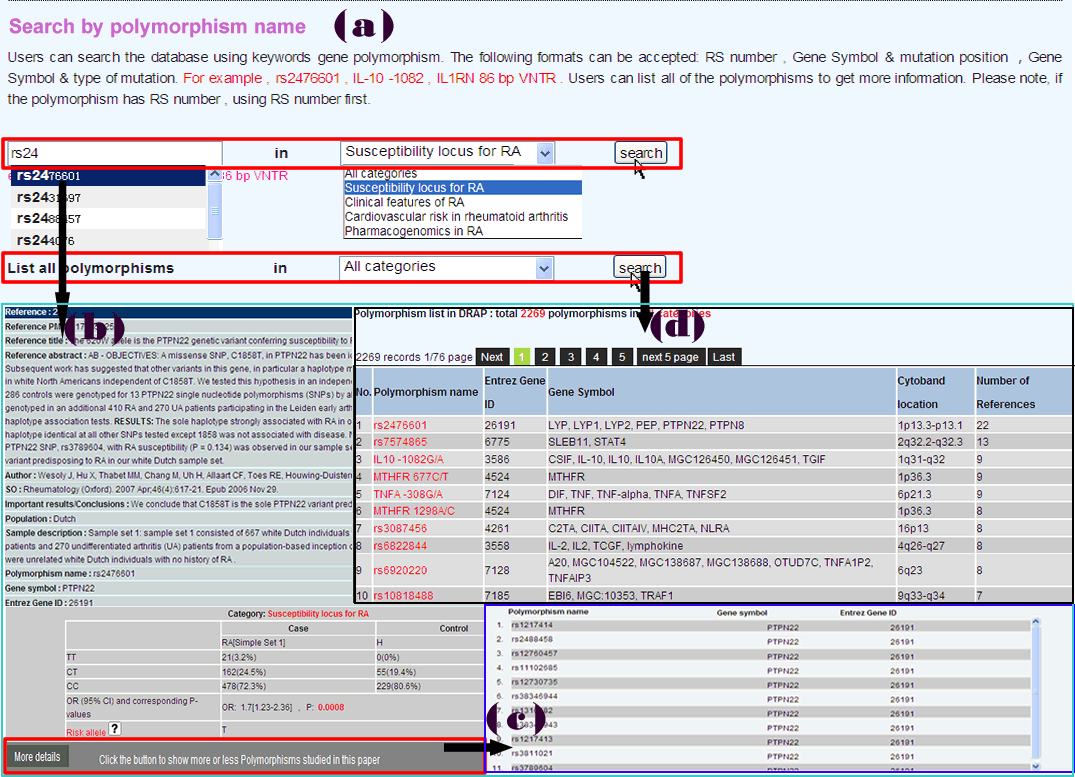
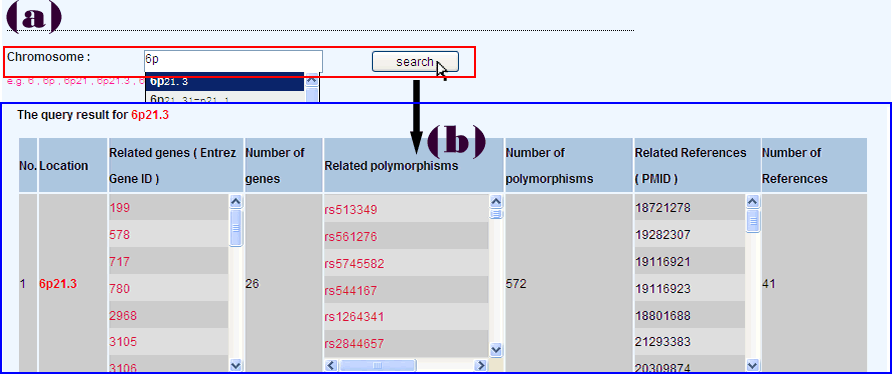
Searching RADB by polymorphism name is a basic function (Figure 1a). There are several types of polymorphisms in RADB, such as single nucleotide polymorphism, HLA allele, microsatellite. Users can use the dbSNP “rs” number, gene symbol plus mutation position, or gene symbol plus type of mutation to query RADB: for instance “rs2476601”, “PTPN22 1858C/T”, “HLA-DRB1*0401” or “IL1RN 86 bp VNTR”. As mentioned in data categories, data is divided into four categories. Users can choose a category of interest or all categories in this step. If users have no idea about query, they can list all of the polymorphisms in RADB (Figure 1d) . To facilitate the users , an autocomplete function has been used.
The result page will be displayed in a new page (Figure 1b). It is reference centered. That is, each record is a record of a reference. If a polymorphism was investigated in 10 references, there will be 10 records. Query results include basic information about the articles (e.g. PMID, title, Source and important results/conclusions), population information (e.g. population, population details and sample description) and polymorphism information (e.g. polymorphism name, gene symbol, Entrez Gene ID, genotype, OR (95% CI), P-value and risk allele). If an article examined other polymorphisms, a button will appear at the bottom of each record, users can click this button to display more polymorphisms studied in the same paper (Figure 1c).
4.2 Search by gene name
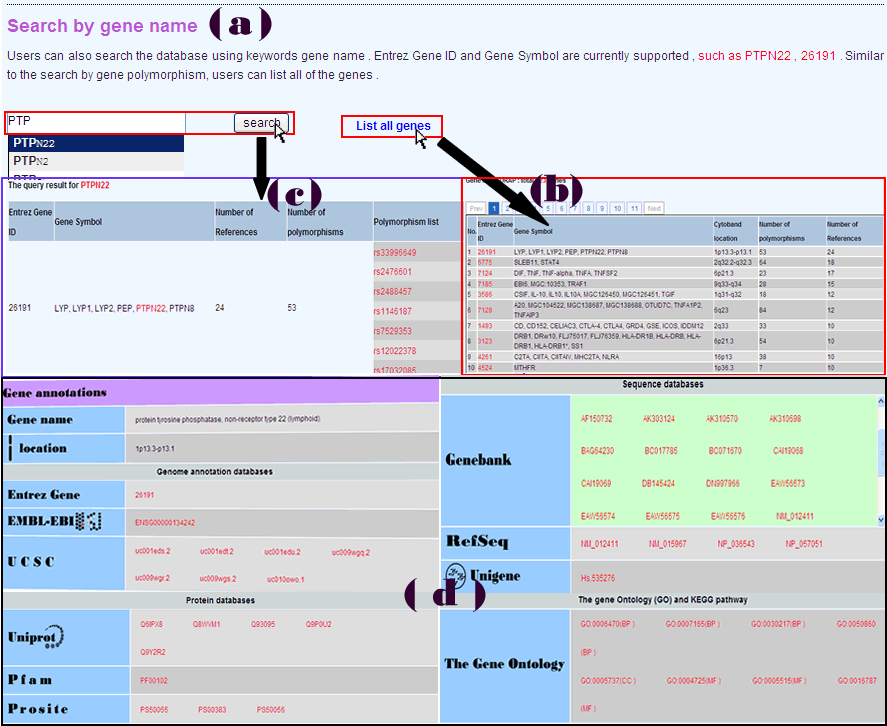
Users can query the database using keywords gene name (Figure 2a) or list all of the genes in RADB (Figure 2b). Both Entrez Gene ID and Gene Symbol are currently supported, (e.g., PTPN22, 26191). The result page will be displayed in a new page (Figure 2c). Query result includes gene-related information in RADB (e.g. Number of References, Number of polymorphisms, Polymorphism list) and hyperlinked gene annotations (e.g. Gene name, Location, Entrez Gene, EMBL-EBI, USCS, Genebank, RefSeq, Unigene, Uniprot, Pfam, Prosite, GO, KEGG pathway) (Figure 1d).
4.3 Search by population
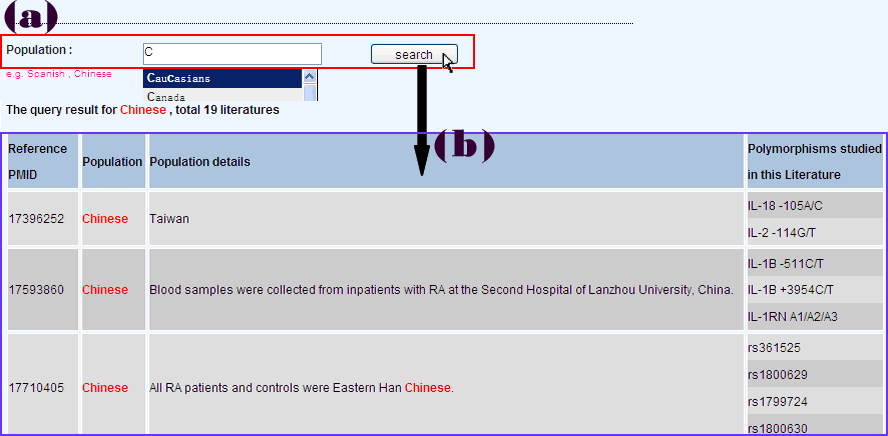
The allelic frequencies of genes often differ substantially between populations, and thus, ethic-specific association studies often get people's attention. Therefore, we provide a query mode of population (Figure 3a). Query result will list all literatures with the same study population and polymorphisms studied in the corresponding literature (Figure 3b).
4.4 Search by different types of research
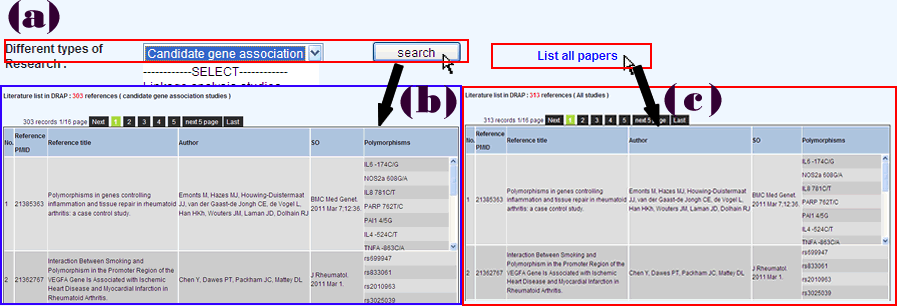
The literatures are divided into three types: candidate gene linkage analysis studies, candidate gene association studies and genome-wide association studies(GWAS).Users can query one of them (Figure 4a) or list all of the literatures (Figure 4c). Query result will list all literatures with the same study type and polymorphisms studied in the corresponding literature (Figure 4b).
4.5 Search by chromosome
Users can search RADB by chromosome (such as “6”, “6p”, “6p21”, “6p21.3”, or “6p21.3–p21.2”) (Figure 5a). The results will list all of the genes and their corresponding polymorphisms located in the queried chromosome or chromosomal region. (Figure 5b).
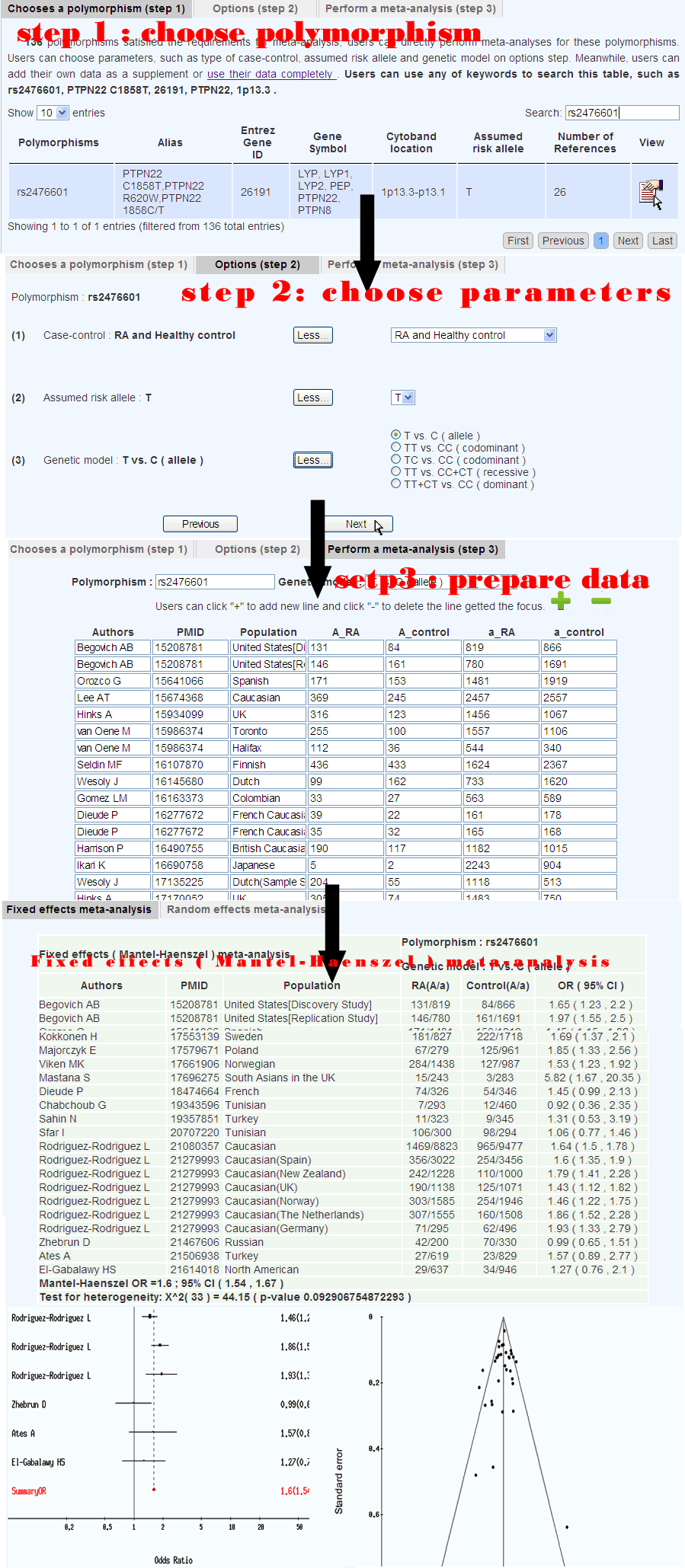
5. Meta-analysis
| Study i | Allele A | Allele a |
|---|---|---|
| RA | ai | bi |
| Control | ci | di |
The results of different association studies often show inconsistencies. A comprehensive evaluation of these results is important. Thus, we developed a module to perform a direct meta-analysis on the polymorphisms in RADB (Figure 6). Users can choose the parameters, such as the type of study (e.g., case-control), the assumed risk allele, and the genetic model. In addition, users can either analyze just their own data or supplement it with RADB data. In our meta-analysis module, the OR and 95% CI are calculated to assess the strength of association. Statistical heterogeneity among the studies is assessed with Woolf's test . A fixed-effects model using the Mantel-Haenszel method and the random effects model of DerSimonian and Laird are used to summarize the results. The summary results are presented in tabular form and forest plots. We also provide a funnel plot to detect publication biases. Warning: where zeros cause problems with computation of effects or standard errors, 0.5 is added to all cells for that study, except when ai=ci=0 or bi=di=0 , when the relative effect measures OR are undefined (ommit this study).
6. Download
To continually improve our database, we welcome the ongoing submission of new data. The submission process is very simple. Users are only required to submit the article’s PMID and the corresponding polymorphism names. We will verify and input the data, if they meet our requirements, as soon as possible ( Figure 7 ).

7. Polymorphism and gene annotation
To obtain more information, we added hyperlinks to external databases: dbSNP or the IMGT/HLA database for polymorphisms; and the NCBI Gene, EMBL-EBI, sequence databases (NCBI GenBank, RefSeq, and Unigene), protein databases (Uniprot, Pfam, and Prosite), and biological pathway databases (GO, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway) for genes. First, we converted gene symbol in the literature to Entrez Gene ID, then we used Entrez Gene ID as the central ID for cross linking and annotation. An R/BioConductor package, named org.Hs.eg.db, was used to map Entrez Gene ID to genome annotation databases mentioned above.