
Statistics

1 . Different types of Research
| CGAa | GWAb | CGLAc | Total |
|---|---|---|---|
| 640 | 25 | 21 | 686 |
aCGA : candidate gene association studies
bGWA : genome-wide association studies
cCGLA : candidate gene linkage analysis studies
2 . The statistical information of polymorphisms
| SNPa | HLA alleleb | minisatellitec or microsatellited | others |
|---|---|---|---|
| 2933 | 175 | 72 | 55 |
aSNP : Single nucleotide polymorphism
bHLA : human leukocyte antigen
cminisatellite : variable number of tandem repeats
dmicrosatellite : short tandem repeat,STR,SSR
3 . Hotspots for rheumatoid arthritis
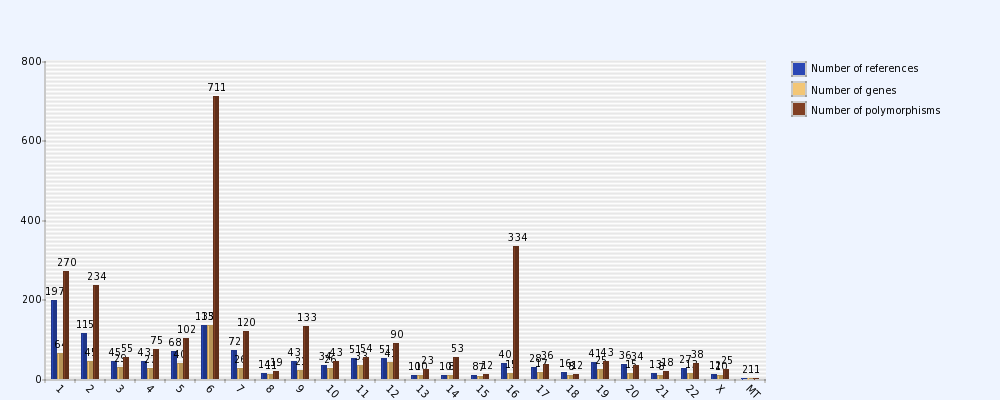
Fig.1. Distribution of rheumatoid arthritis (RA) related polymorphisms , genes and references on chromosomes in RADB. 1-22 represents 22 pairs of autosomes, ‘X’ represents X chromosome. The Y-axis indicates number of polymorphisms , genes or references.
We can clearly see that polymorphisms mainly distribute on chromosome 6, chromosome 1, chromosome 2 , chromosome 9 and chromosome 16 . The distribution of genes is similar to the distribution of polymorphisms. There are more genes on chromosome 1 , chromosome 2 and chromosome 6 than others.
Table 1. The statistical information of top 50 cytoband locations in RADB (sorted by number of references)
4 . Top 50 genes of positive
Table 2. The statistical information of top 50 genes in RADB (sorted by number of statistically significant references)
| No. | Entrez Gene ID | Gene Symbol | Chromosome | Startloci | Endloci |
|
||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 26191 | PTPN22 | 1 | 114356433 | 114414375 | 50 | 37 |
| 2 | 7124 | TNF | 6 | 31543344 | 31546113 | 41 | 23 |
| 3 | 6775 | STAT4 | 2 | 191894302 | 192016322 | 29 | 21 |
| 4 | 1493 | CTLA4 | 2 | 204732511 | 204738683 | 33 | 17 |
| 5 | 23569 | PADI4 | 1 | 17634690 | 17690495 | 25 | 16 |
| 6 | 3586 | IL10 | 1 | 206940948 | 206945839 | 20 | 10 |
| 7 | 2214 | FCGR3A | 1 | 161511549 | 161520413 | 12 | 8 |
| 8 | 3553 | IL1B | 2 | 113587337 | 113594356 | 17 | 8 |
| 9 | 7185 | TRAF1 | 9 | 123664671 | 123691451 | 17 | 8 |
| 10 | 115352 | FCRL3 | 1 | 157647978 | 157670647 | 15 | 8 |
| 11 | 3569 | IL6 | 7 | 22766766 | 22771621 | 16 | 8 |
| 12 | 79258 | MMEL1 | 1 | 2522081 | 2564481 | 13 | 7 |
| 13 | 4524 | MTHFR | 1 | 11845787 | 11866160 | 14 | 7 |
| 14 | 3663 | IRF5 | 7 | 128577994 | 128590089 | 9 | 7 |
| 15 | 3565 | IL4 | 5 | 132009678 | 132018370 | 12 | 7 |
| 16 | 7128 | TNFAIP3 | 6 | 138188325 | 138204451 | 9 | 6 |
| 17 | 4855 | NOTCH4 | 6 | 32162620 | 32191844 | 6 | 6 |
| 18 | 10665 | C6orf10 | 6 | 32260475 | 32339656 | 5 | 5 |
| 19 | 56244 | BTNL2 | 6 | 32362513 | 32374900 | 6 | 5 |
| 20 | 23534 | TNPO3 | 7 | 128594234 | 128695227 | 9 | 5 |
| 21 | 3606 | IL18 | 11 | 112013974 | 112034840 | 9 | 5 |
| 22 | 1235 | CCR6 | 6 | 167525295 | 167552629 | 8 | 5 |
| 23 | 7040 | TGFB1 | 19 | 41836812 | 41859831 | 12 | 5 |
| 24 | 4049 | LTA | 6 | 31539876 | 31542101 | 7 | 5 |
| 25 | 958 | CD40 | 20 | 44746906 | 44758384 | 14 | 5 |
| 26 | 5966 | REL | 2 | 61108752 | 61150178 | 9 | 5 |
| 27 | 399716 | DKFZp667F0711 | 10 | 6392283 | 6392831 | 10 | 4 |
| 28 | 84162 | KIAA1109 | 4 | 123091758 | 123283914 | 6 | 4 |
| 29 | 3566 | IL4R | 16 | 27325230 | 27376099 | 6 | 4 |
| 30 | 7133 | TNFRSF1B | 1 | 12227060 | 12269277 | 11 | 4 |
| 31 | 6891 | TAP2 | 6 | 32789610 | 32806547 | 7 | 4 |
| 32 | 5788 | PTPRC | 1 | 198608098 | 198726605 | 7 | 4 |
| 33 | 4795 | NFKBIL1 | 6 | 31514628 | 31526606 | 6 | 4 |
| 34 | 5243 | ABCB1 | 7 | 87133179 | 87342639 | 5 | 4 |
| 35 | 26147 | PHF19 | 9 | 123617931 | 123639606 | 5 | 4 |
| 36 | 3559 | IL2RA | 10 | 6052657 | 6104333 | 8 | 4 |
| 37 | 10666 | CD226 | 18 | 67530192 | 67624232 | 6 | 4 |
| 38 | 7189 | TRAF6 | 11 | 36505317 | 36531863 | 7 | 4 |
| 39 | 4314 | MMP3 | 11 | 102706528 | 102714342 | 5 | 4 |
| 40 | 150577 | LINC01104 | 2 | 100824716 | 100867946 | 6 | 4 |
| 41 | 7919 | DDX39B | 6 | 31497996 | 31510252 | 4 | 4 |
| 42 | 7421 | VDR | 12 | 48235320 | 48298814 | 7 | 4 |
| 43 | 199 | AIF1 | 6 | 31582994 | 31584798 | 4 | 4 |
| 44 | 90355 | C5orf30 | 5 | 102594442 | 102614361 | 5 | 3 |
| 45 | 4261 | CIITA | 16 | 10971055 | 11018840 | 13 | 3 |
| 46 | 84868 | HAVCR2 | 5 | 156512843 | 156536248 | 3 | 3 |
| 47 | 23274 | CLEC16A | 16 | 11038345 | 11276046 | 5 | 3 |
| 48 | 9374 | PPT2 | 6 | 32121229 | 32131458 | 3 | 3 |
| 49 | 54665 | RSBN1 | 1 | 114304454 | 114355070 | 5 | 3 |
| 50 | 326 | AIRE | 21 | 45705721 | 45718102 | 3 | 3 |
a Stat. Significant : P value < 0.05 was considered significant.